
 |
1) Essential enzymes of human pathogens
|
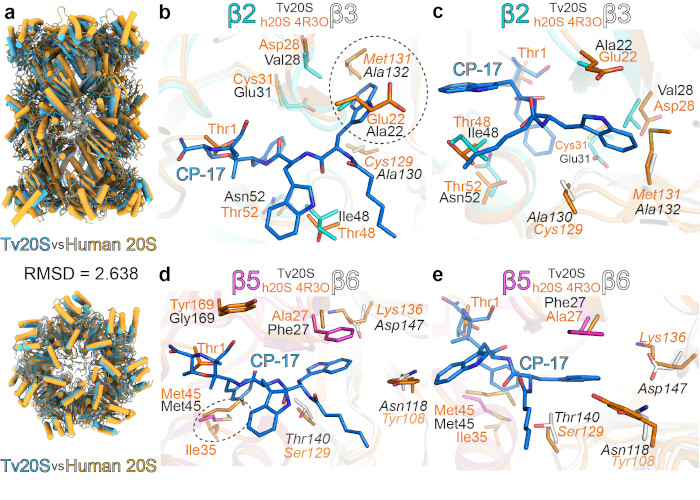
Every pathogenic microorganism has several key enzymes that are essential for its replication, and inhibitors of these enzymes therefore represent potential drugs. Most of the drugs developed at the IOCB (such as tenofovir) are antivirals. This tradition continues; however, we are also focusing on other pathogenic microorganisms. Shown below is the proteasome from the highly pathogenic protist Trichomonas vaginalis. |
||
 |
||
Structural comparison of Trichomonas vaginalis and human proteasomes (Silhan et al., Nature Comm. 2024). |
||
2) Viral hijacking of human host factors
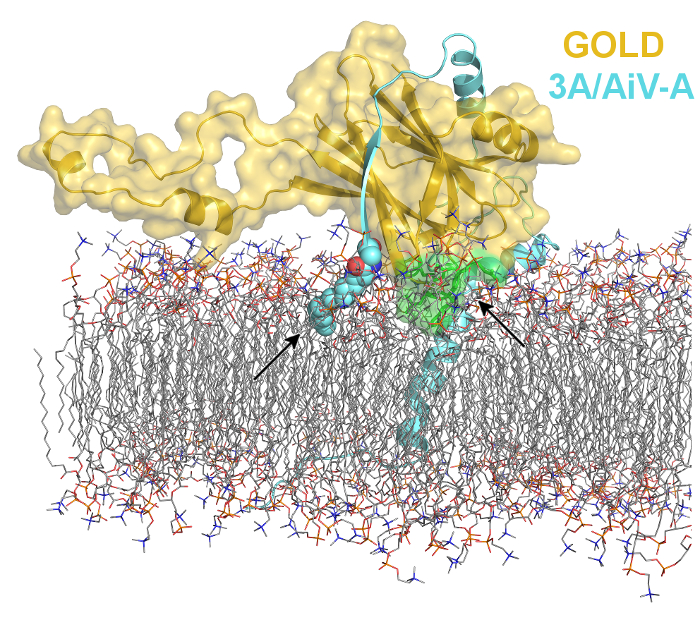
Viruses are way too small organism (not even considered living by many) to encode all the proteins they require to reproduce in our cells. Therefore, they evolved mechanisms how to hijack our proteins for their selfish benefits. We use protein crystallography to understand molecular principles of viral reproduction in our cells. Our research not only reveals the mysteries from viral life cycle but also provides new targets for drug design. |
||
 |
||
Viral non-structural 3A proteins act as a molecular lasso to hijack the Golgi resident ACBD3 protein (Klima et al. 2017). Molecular dynamics simulation-based model of the ACBD3 GOLD domain in complex with viral 3A protein based on our recent crystal structure (Klima et al. 2017) on the lipid bilayer. The viral 3A protein is colored in aquamarine and shown in cartoon representation except for the myristoylated G1 residue which is shown in spheres representation and colored according to elements - carbons are colored in aquamarine, oxygens in red, nitrogen in blue. The ACBD3 GOLD domain is shown in cartoon representation with semi-transparent surface and colored in gold except for the membrane-binding site composed of R399, L514, W515, and R516, which is colored in green. |
||
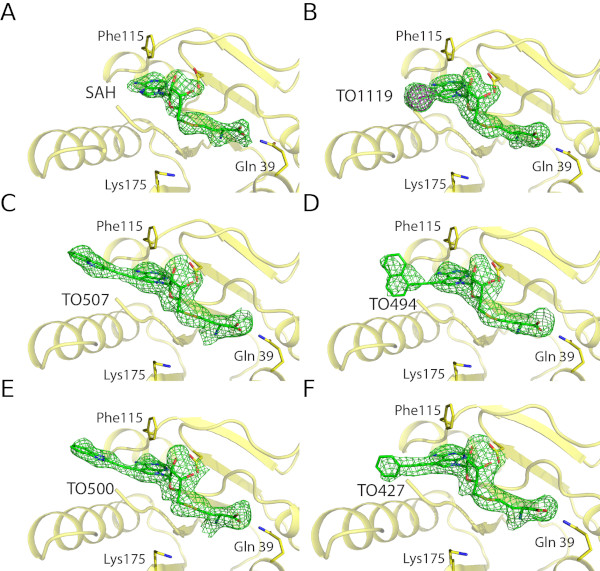
3) Structure guided inhibitor design
Structural analysis of inhibitors in the active site of a viral enzyme (Silhan et al., Nature Comm. 2023). |
||
 |
||
| Inhibitors ot the monkey pox virus methyltransferase VP39 synthezied at the group of Dr. Nencka (IOCB) in the enzyme's active site. Electron density of the inhibitors shown as green mash. | ||
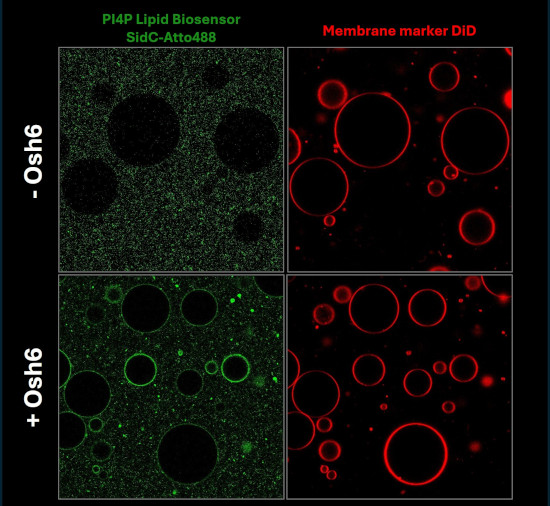
4) Lipid transport proteins
Visualization of lipid transport Lipids are not only transported via small lipid vesicles but also by lipid transport proteins. We aim to biophysically characterize this process and to visualise the lipid transport in vitro using advancend lipid biosensors.
|
||
 |
||
| PI4P-containing Large Unilamellar Vesicles (LUVs) were added to phosphatidylserine-containing Giant Unilamellar Vesicles (GUVs). The fluorescently labeled lipid biosensor SidC was used as a PI4P marker. Upon addition of the lipid transport protein Osh6, SidC redistributed from the LUVs to the GUVs, indicating transfer of PI4P between the membranes. | ||