Research from IOCB Prague reveals a previously unknown mechanism of genetic transcription

Scientists at IOCB Prague are uncovering new details of gene transcription. They have identified a previously unknown molecular mechanism by which the transcription of genetic information from deoxyribonucleic acid (DNA) into ribonucleic acid (RNA) can be initiated. The researchers focused on a specific class of molecules known as alarmones, which are found in cells across a wide range of organisms and whose levels often increase under conditions of cellular stress. The results were published in the prestigious scientific journal Nature Chemical Biology.
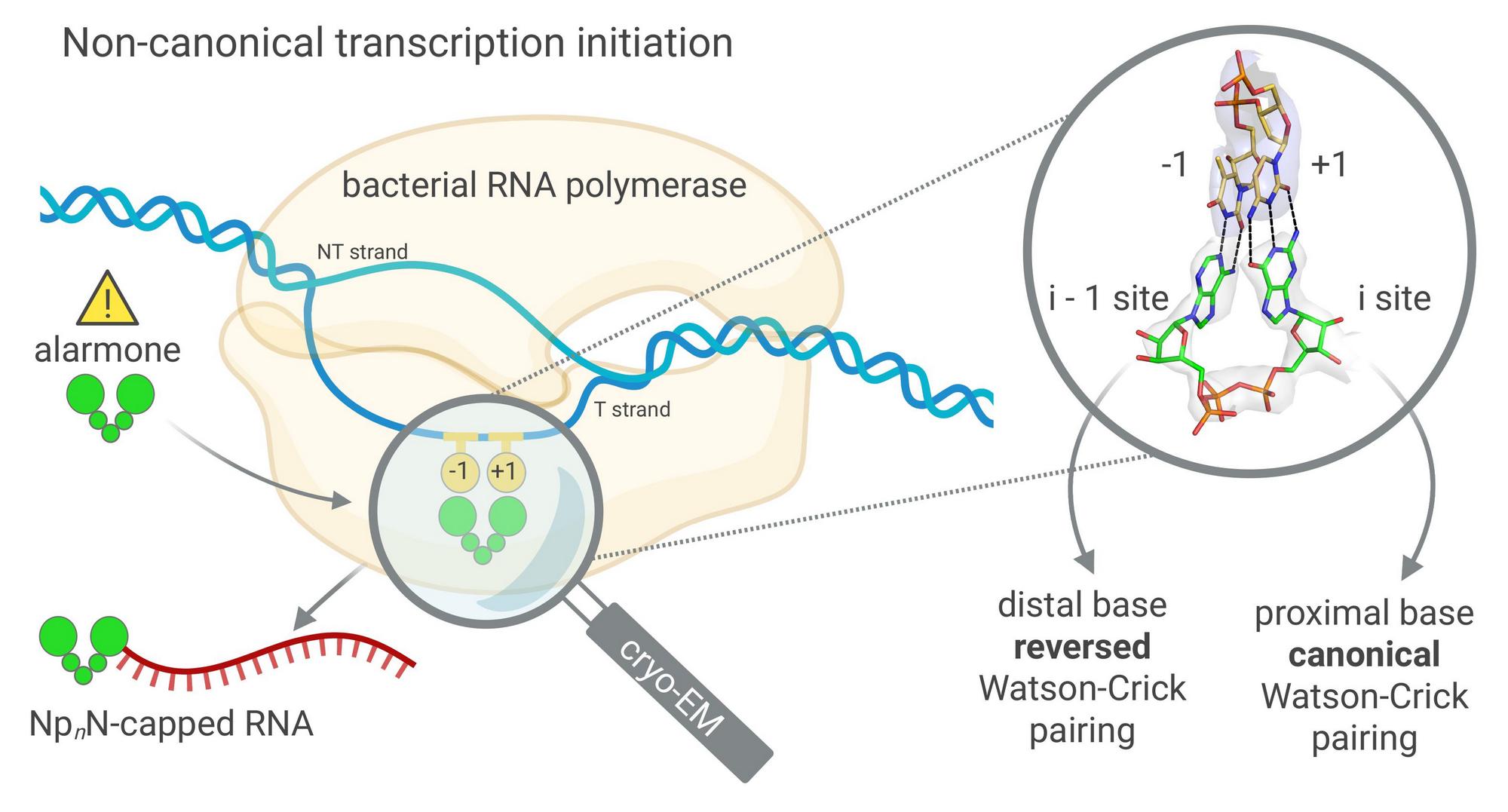
Molecules of ribonucleic acid can carry a variety of chemical modifications at one end, known as caps. In eukaryotic organisms, including human cells, the best-known cap plays an important role in RNA stability and in regulating its subsequent fate. In recent years, however, it has become clear that alternative, non-canonical RNA caps also exist, although their formation and the mechanisms by which they are attached to RNA remain only partially understood. These include alarmone caps, formed by dinucleoside polyphosphate molecules, that protect cellular RNA at moments when the cell is under threat.
In their study, Dr. Hana Cahová and colleagues examined bacterial RNA polymerase and investigated how this enzyme can initiate transcription using dinucleoside polyphosphates (NpNs) instead of the standard RNA building blocks. For the first time, the scientists described, at the atomic level, how RNA bearing an alarmone cap can be generated directly at the initiation of gene transcription. They also observed that NpNs bind through a different type of base pairing than is typically observed.
The work was significantly shaped by Valentina Serianni from Hana Cahová’s team, who demonstrated the ability of dinucleoside polyphosphates to initiate gene transcription, and by Jana Škerlová, who focused on the structural analysis of RNA polymerase. Using cryogenic electron microscopy data, she showed how dinucleoside polyphosphate molecules bind within the active site of RNA polymerase – that is, the core of the enzyme where genetic information is transcribed. Together, their findings help to elucidate the processes that accompany the transcription of genetic information from DNA to RNA.
Hana Cahová adds: “We’re describing something that truly occurs in cells and that we’re now able to observe directly at the level of individual molecules. This allows us to answer fundamental questions about cellular processes, such as how cells adapt to stress. RNA plays a central role in this, as it carries the cascade of information underlying any cellular response – for example, to threatening conditions caused by nutrient deprivation or temperature shock.”

Cryogenic electron microscopy (cryo-EM) was central to the project. This part of the work was led by Dr. Tomáš Kouba, one of the authors of the study. “Cryogenic electron microscopy allows us to freeze biological molecules in a state very close to their natural form and then determine their three-dimensional structure. This makes it possible to look directly into the active centers of enzymes and observe their function down to the atomic level,” he explains.
Last year, IOCB Prague opened a new cryo-EM center that is unique within the Czech research environment. State-of-the-art cryogenic electron microscopes are housed in a specially designed building and enable the study of biological processes with exceptional structural precision.
Original publication
- Serianni, V. M.; Škerlová, J.; Dubánková, A. K.; Škríba, A.; Šváchová, H.; Vučková, T.; Filimoněnko, A.; Fábry, M.; Řezáčová, P.; Kouba, T.; Cahova, H. Molecular insight into 5′ RNA capping with NpnNs by bacterial RNA polymerase. Nat. Chem. Biol. 2026. https://doi.org/10.1038/s41589-025-02134-5