Natural product-inspired macrocycles – Concert of theory and synthesis toward biological activity
PI: Ullrich Jahn, co-PI: Lubomír Rulíšek
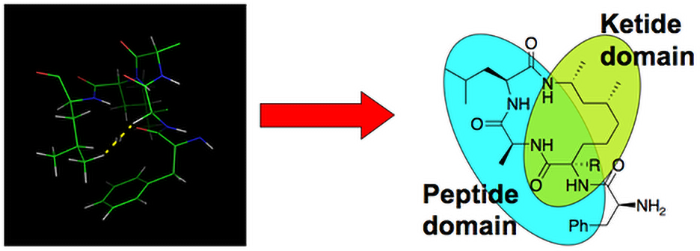
Nature provides us with highly biologically active macrocycles, which are widely used as drugs for many indications. In contrast, macrocycles have so far not played a dominant role in conventional drug design, probably because of difficulties to control conformation and consequently proper and strong binding to receptors on one hand and difficult syntheses on the other hand.
In our project we develop new combined computational‐synthetic strategies for the design of strongly binding macrocycles. The approach is based on the computational power to map the active sites of receptors, conformational space of macrocycles and to identify key binding domains in silico. Based on this three‐dimensional conformationally flexible macrocyclic compounds with small strain energies are designed that connect the binding domains and can adapt to the space provided by the receptors. An attractive, but so far unused feature is the strategic introduction of structural elements that Nature uses successfully to optimize interactions by limiting the number of conformations by motives present in peptides, polyketides or terpenes.
This principle may be applicable in many therapeutic fields for the design of lead compounds ranging from antiviral to anticancer. For example, as potentially efficient binders of the C-terminal domain of the HIV-1 capsid, several polyketide‐peptide macrocycles were identified and successfully synthesized. Syntheses of other polyketide‐peptide macrocycles inspired by peptide nucleic acids as potential ligands of the stimulator of interferon genes (STING) protein are ongoing.



