Small-molecule inhibitors of viral replication
PI: Radim Nencka, co-PIs: Evžen Bouřa, Václav Veverka
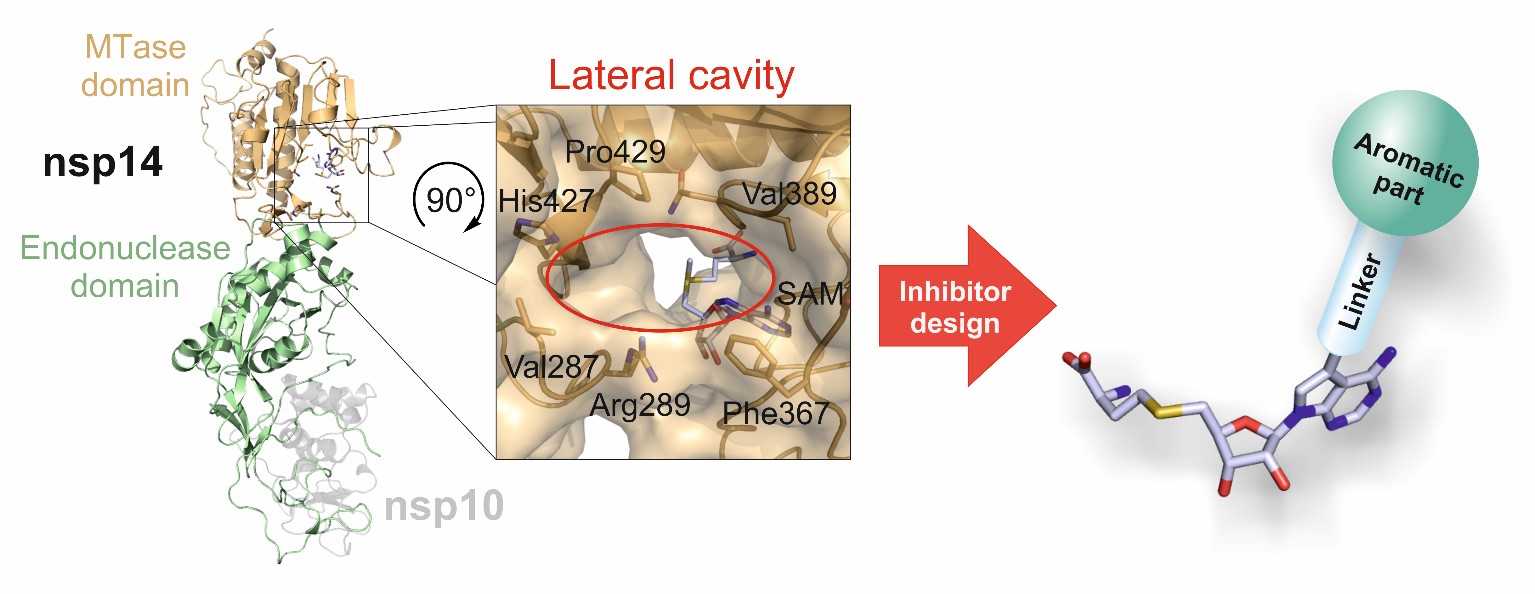
The main goal of this project is the rational design of new antiviral compounds. Our method of designing new compounds is based on crystal structures of target proteins and the identification of small fragments by NMR spectroscopy. The actual positions of the rationally designed compounds and fragments are determined by X-ray crystallography or NMR spectroscopy and/or predicted by molecular docking. Based on this structural information, we then attempt to further refine and "grow" the compounds into high-affinity ligands. In the recent past, we have used these methodologies to identify suitable ligands for key viral enzymes from several important groups of human pathogens, in particular viruses from Flaviviridae family.
Recently, however, the COVID-19 pandemic has prompted us to focus significantly on individual coronavirus representatives as well, and SARS-CoV-2 and in particular its methyltransferases and polymerase, which are essential components of its replication apparatus, have become the focus of our interest. This program combines aspects of several scientific disciplines, including medicinal chemistry, biochemistry, structural and molecular biology, and molecular docking. It leverages the unique facilities available to team members at IOCB for drug discovery projects, which can be mutually beneficial to IOCB and Gilead Sciences.



