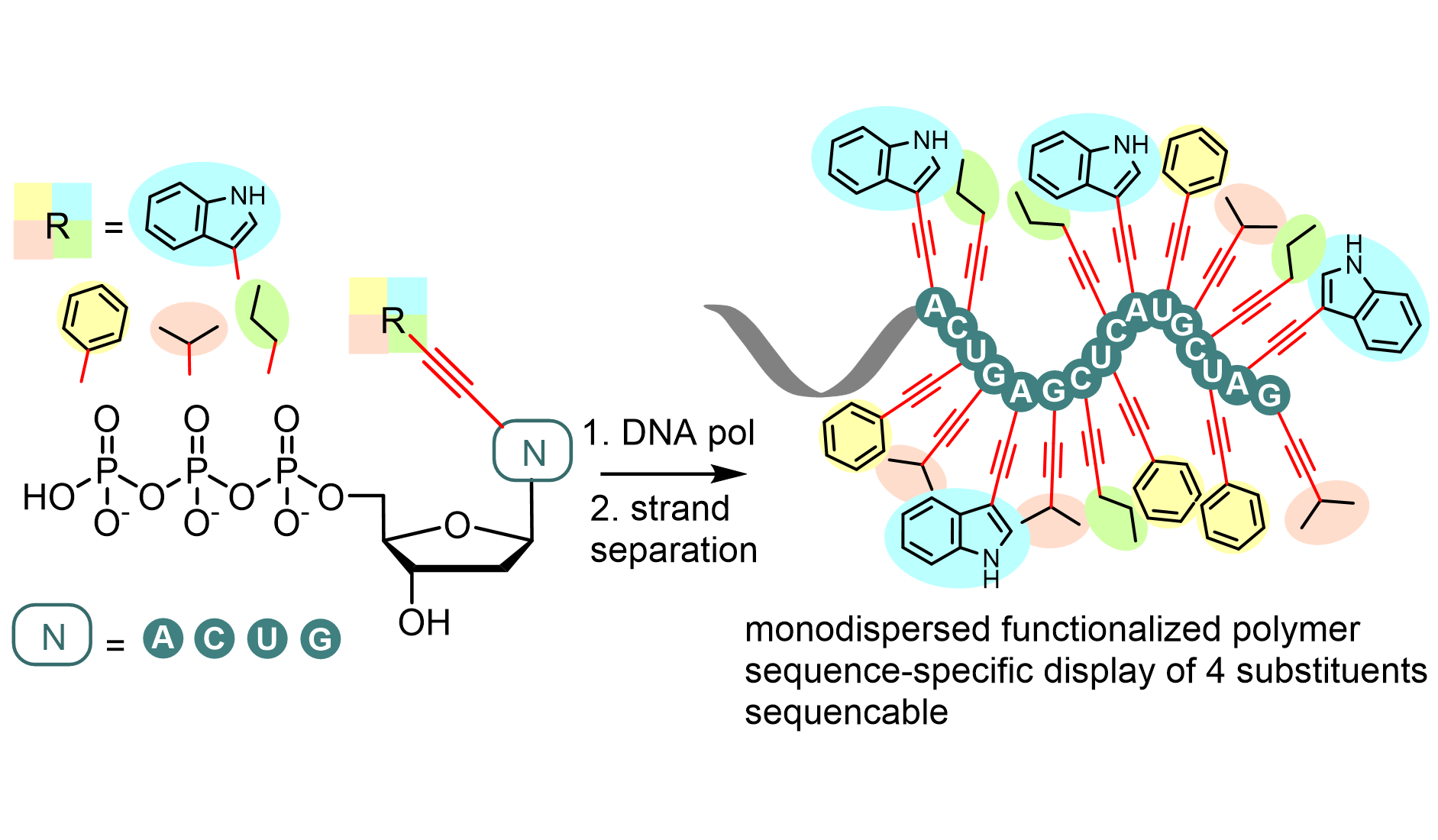
A team of researchers, led by Michal Hocek with Marek Ondruš as the first author, described in the journal Nucleic Acids Research the development of an approach for the enzymatic synthesis of hypermodified DNA, where all the canonical nucleotides are replaced by modified counterparts equipped with four different hydrophobic chains similar to side-chains of amino acids. The resulting polymer is based on a DNA scaffold which presents small molecule substituents in a sequence specific way.
High fidelity of the incorporation of the modified dNTPs was confirmed by the next-generation sequencing of long sequences after converting the modified DNA back to the natural unmodified DNA. Biophysical analyses revealed that such heavily hydrophobic-modified DNA also adopted the most abundant B-DNA structure and had hybridization properties comparable with unmodified DNA.
These results are particularly useful for those in the community who are interested in the scope of DNA polymerases and biotech applications. This approach is now being used in the Hocek group Hocek group for the generation and selection of hypermodified aptamers for undruggable protein targets.
The original paper: Marek Ondruš, Veronika Sýkorová, Lucie Bednárová, Radek Pohl, Michal Hocek, Enzymatic synthesis of hypermodified DNA polymers for sequence-specific display of four different hydrophobic groups, Nucleic Acids Research, gkaa999, https://doi.org/10.1093/nar/gkaa999