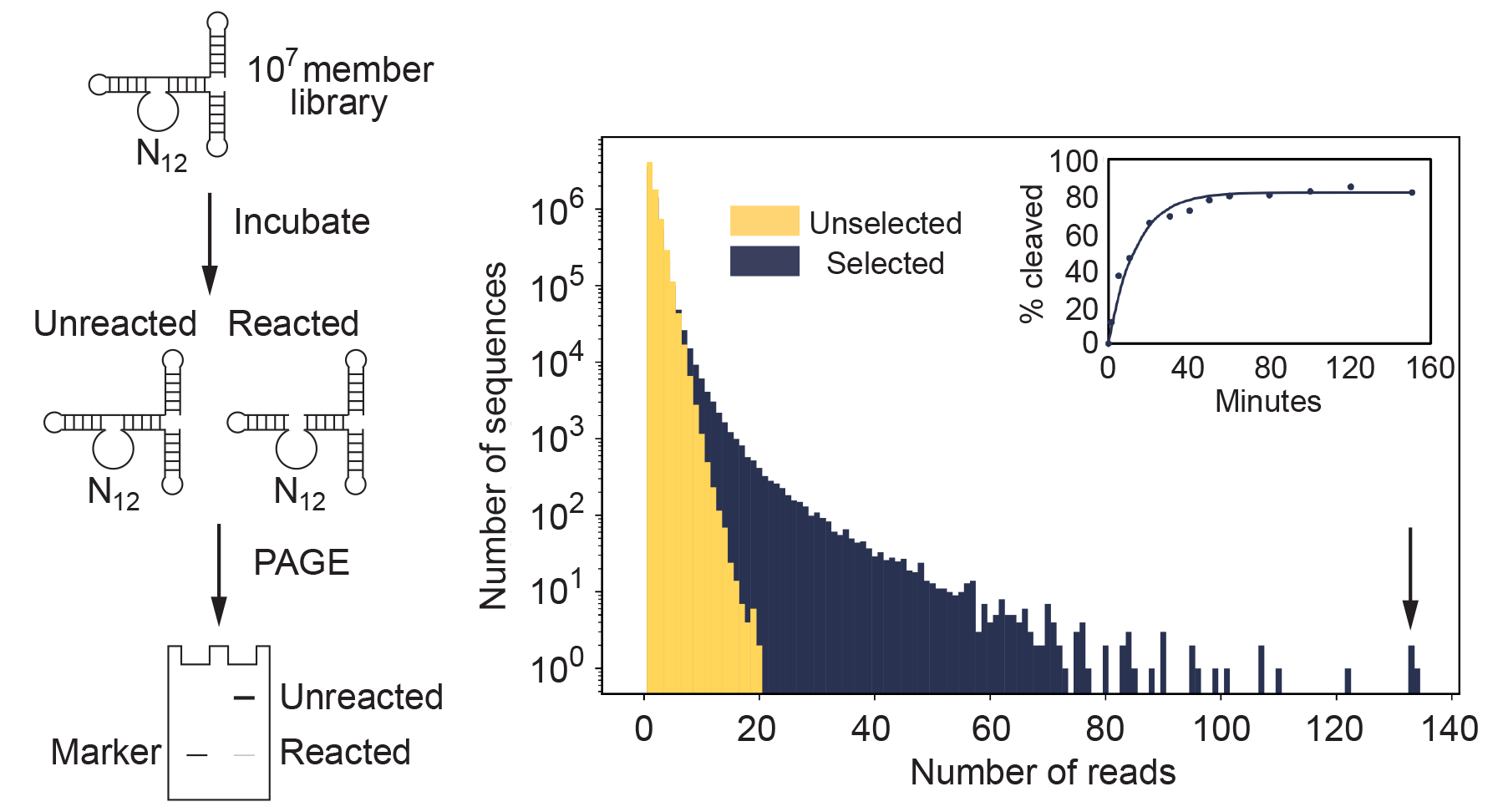
Methods of artificial evolution such as in vitro selection make it possible to isolate DNA and RNA sequences with useful functions from random sequence libraries. Conventional selection experiments use libraries of 1015 sequences and require multiple rounds of selections to identify rare sequences with a desired function.
Based on the simple structures of some functional nucleic acid motifs, researchers in the Curtis group at IOCB Prague hypothesized that it might be possible to identify catalytic DNA molecules (deoxyribozymes) from smaller libraries in a single round of selection followed by high-throughput sequencing.
To test this idea, they embedded 12 or 15 randomized positions (corresponding to 107 to 109 different sequences) in a stable scaffold present in all library members. After a short incubation, sequences that cleaved themselves at an RNA linkage were isolated using a single PAGE purification and characterized by high-throughput sequencing.
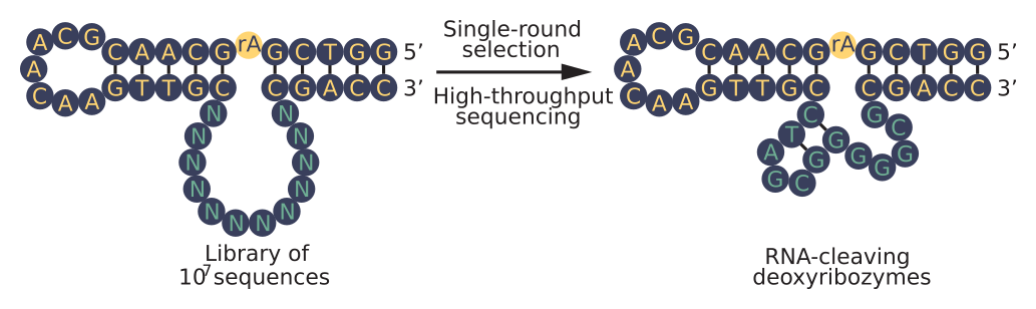
The most active deoxyribozyme identified, named "Dvanactka" after the Czech word for 12, catalyzed RNA cleavage 5000 times faster than the nonenzymatic rate of RNA cleavage under these conditions. Because coverage from high-throughput sequencing was sufficient to detect most enriched sequences, it was also possible to characterize the sequence requirements and secondary structures of some of these motifs without performing additional experiments.
Due to its relative simplicity, we anticipate that this approach will accelerate the discovery of new functional DNA and RNA motifs.
The original article
- Streckerová, T.; Kurfürst, J.; Curtis, E. A. Single-round deoxyribozyme discovery. Nucleic Acids Research 2021. https://doi.org/10.1093/nar/gkab504